Researchers at National Taiwan University have uncovered, for the first time at atomic resolution, how the human proteasome recognizes branched ubiquitin chains. Their finding reveals a multivalent decoding mechanism that enhances protein degradation accuracy and speed.
Protein quality control is central to maintaining cellular homeostasis. Within this system, the ubiquitin–proteasome pathway (UPS) serves as the major mechanism for tagging and degrading unwanted or damaged proteins.
For decades, the prevailing view has been that homotypic K48-linked polyubiquitin chains represent the canonical degradation signal recognized by the proteasome. Yet, despite their textbook status, how these ubiquitin tags are read with molecular precision by the proteasome has remained surprisingly elusive.
Branched ubiquitin chains and proteasome recognition
Emerging evidence has indicated that branched ubiquitin chains, such as those containing mixed K11/K48 linkages, are pivotal in proteostasis, cell cycle progression and stress responses. However, the structural and topological complexity of branched chains—combined with a lack of analytical tools that can simultaneously resolve linkage type, chain length, and three-dimensional conformation—has left a major gap in understanding how these signals are decoded at the molecular level.
A research team led by Dr. Shang-Te Danny Hsu, Research Fellow at the Institute of Biological Chemistry, Academia Sinica, and Adjunct Professor at the Institute of Biochemical Sciences, National Taiwan University, has now overcome this challenge.
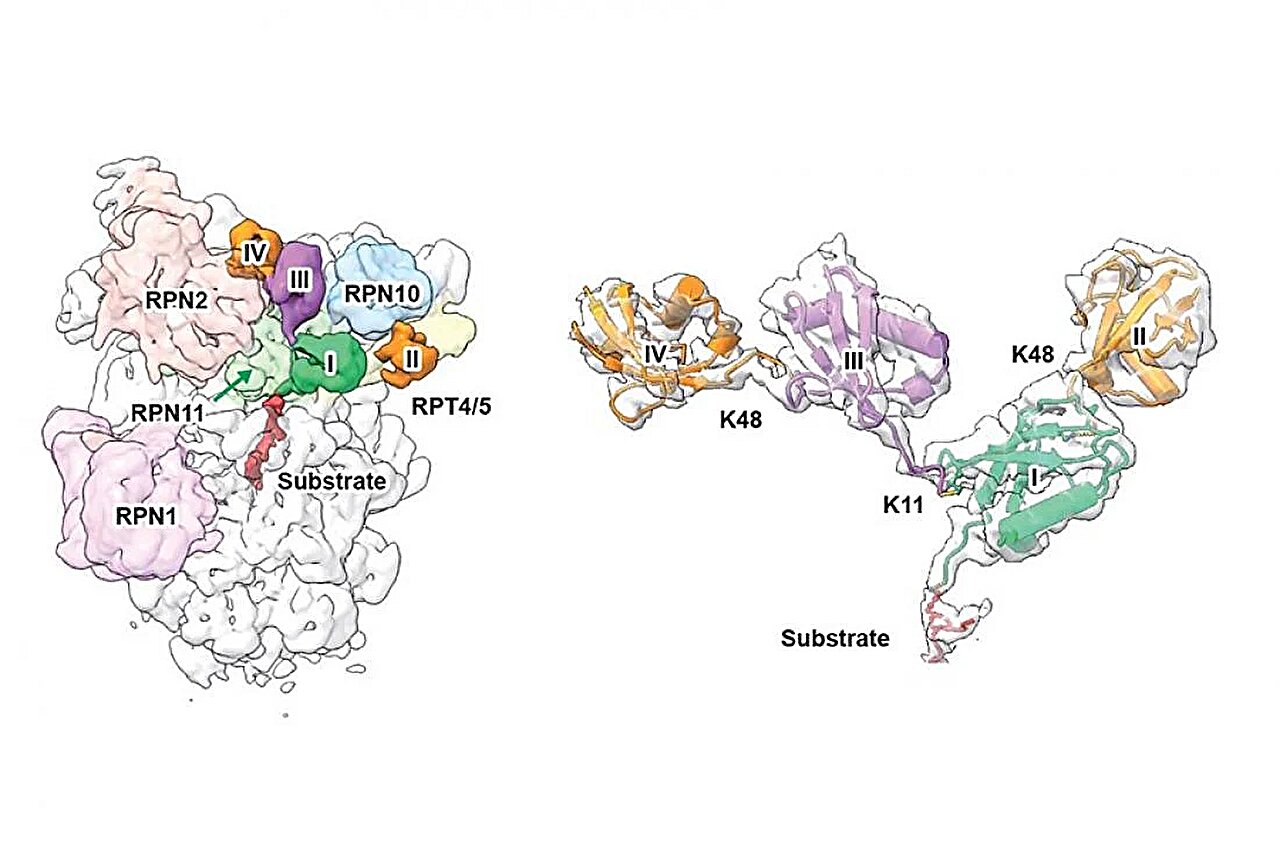
By integrating cryo-electron microscopy (cryo-EM), ubiquitin absolute quantification mass spectrometry (Ub-AQUA), and cross-linking mass spectrometry (XL-MS), the team has successfully determined a high-resolution structure of the human proteasome bound to a branched K11/K48-linked ubiquitin chain. This work precisely defines the branching point and overall topology of the ubiquitin chain, marking the first atomic-level visualization of how the proteasome engages branched ubiquitin architectures.
Key findings and implications for protein degradation
Published in Nature Communications, the study reveals that RPN2, a subunit of the proteasomal 19S regulatory particle, contains a previously unrecognized K11-specific binding site.
Together with other proteasomal subunits including RPN10, RPN8, and RPT4/5, a multivalent recognition groove tailored for K11-linked Ub chains is formed, potentially leading to higher affinity and specificity, thereby enhancing substrate recognition and accelerating degradation. Such a mechanism may equip cells with the ability to rapidly eliminate key regulatory proteins when needed.
"The quantitative power of Ub-AQUA was essential to this discovery. By accurately measuring the abundance of different ubiquitin linkage types, the researchers validated both the chemical composition and the three-dimensional architecture of the assembled branched chains—eliminating ambiguities that commonly arise when relying solely on conventional mass spectrometry or biochemical assays," says Prof. Shang-Te Danny Hsu, corresponding author of the study.
"This integrated 'structure + topology + absolute quantification' strategy was key to resolving a question that has challenged the field for years."
To see article on Phys.org: https://phys.org/news/2025-12-decoding-human-proteasome-ubiquitin-chains.html